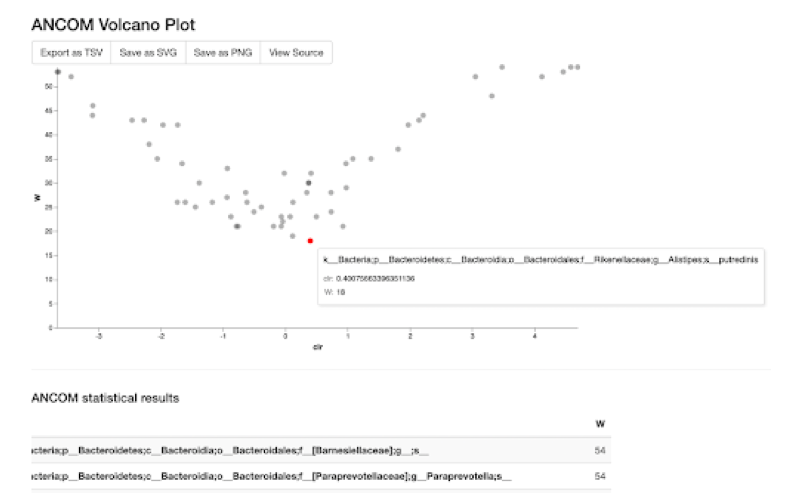
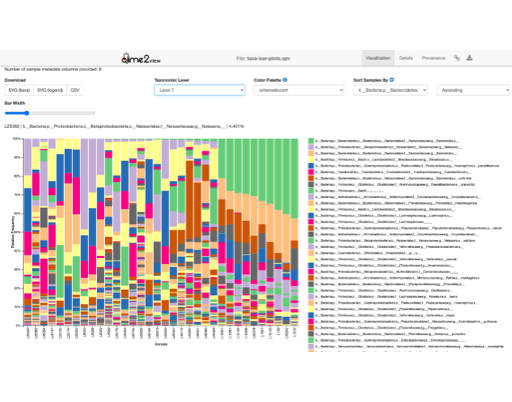
Metagenomics 16S analysis
When using our 16S pipelines, all you would need are your raw reads and the sample metadata. Our pipelines will seamlessly process your data and give you the desired analysis results within no time. With just one click, the output files can be loaded in QIIME2 View, making it easy for you to interactively explore your results and make the breakthrough you've been waiting for.