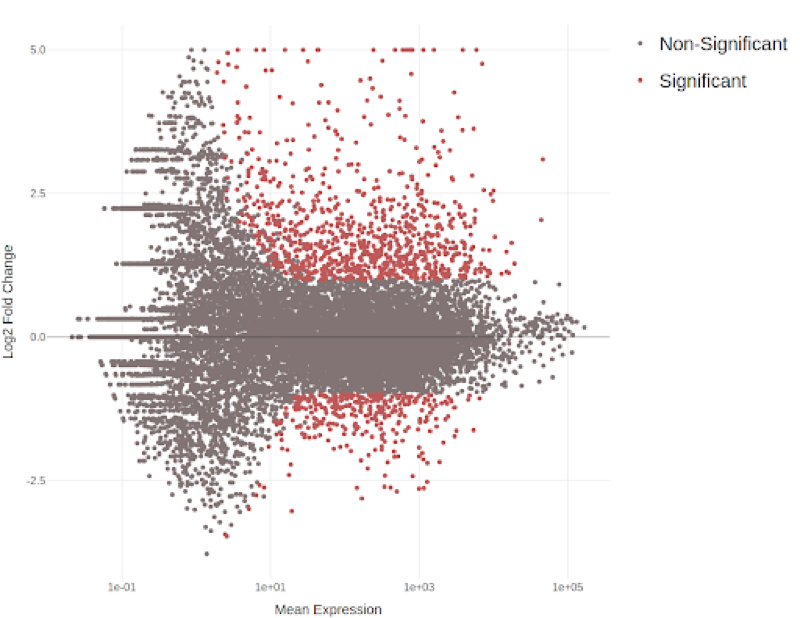
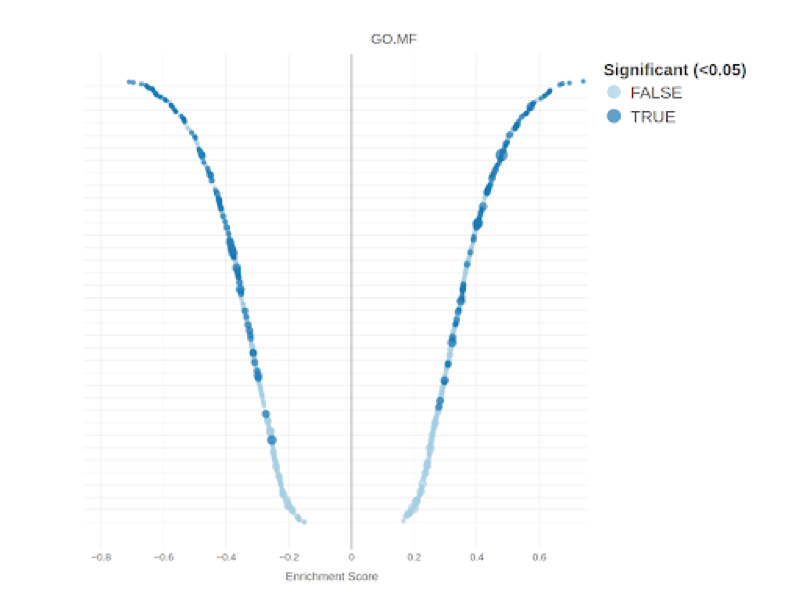
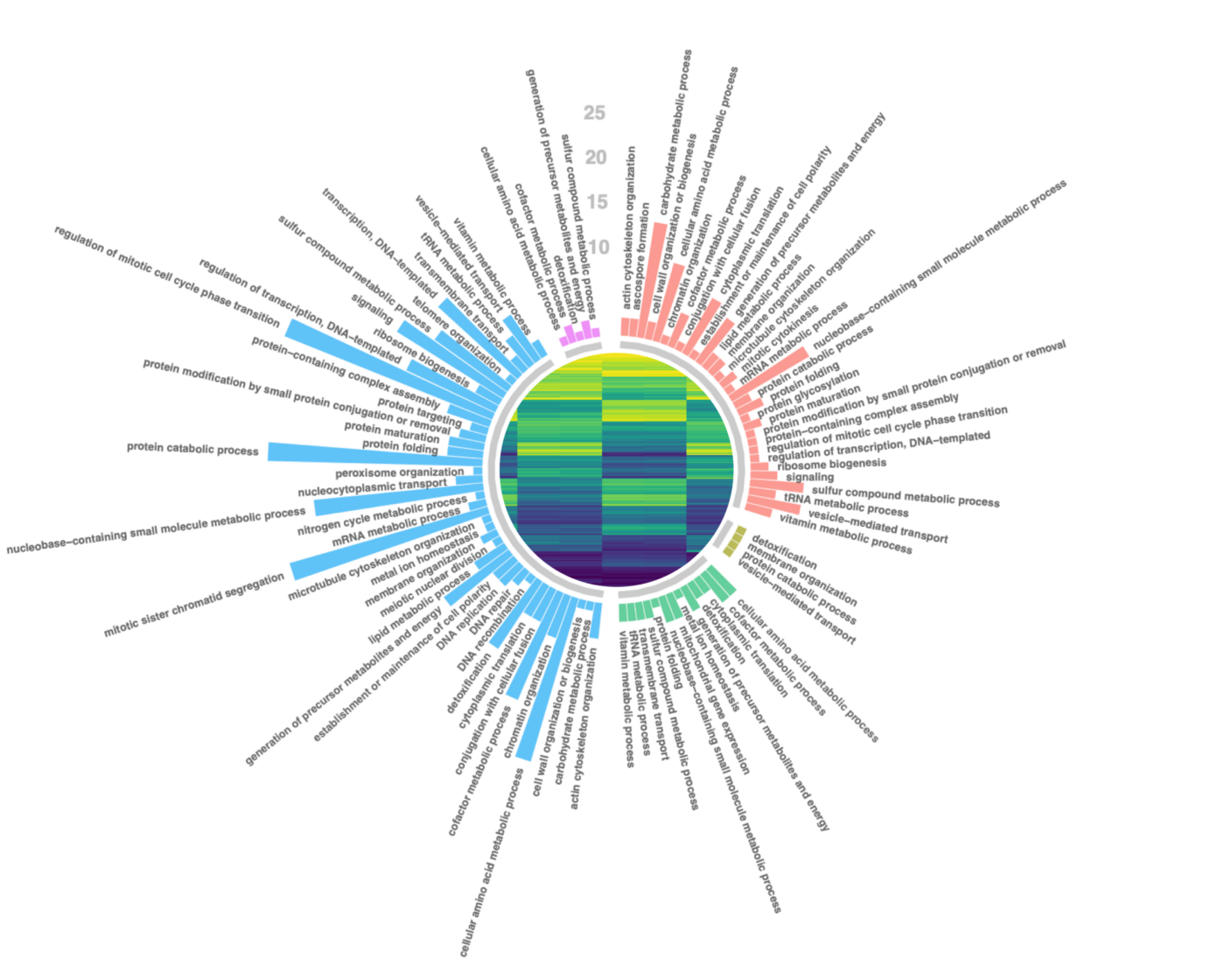
RNA-Seq Expression analysis
RNA Sequencing (RNA-Seq) is a revolutionary technique for transcriptome-wide analysis of gene expression. Given its high accuracy and sensitivity for measuring expression, it has become the standard for studying transcriptomic dynamics and identifying differences in expression between tissues, physiological stages, diseases and a wide range of other experimental designs.