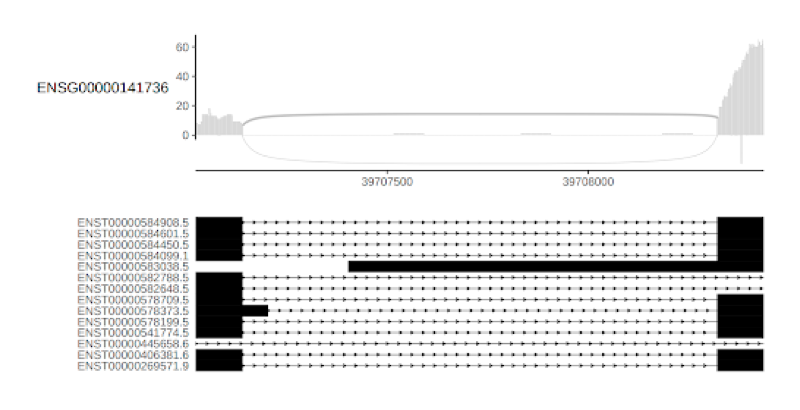
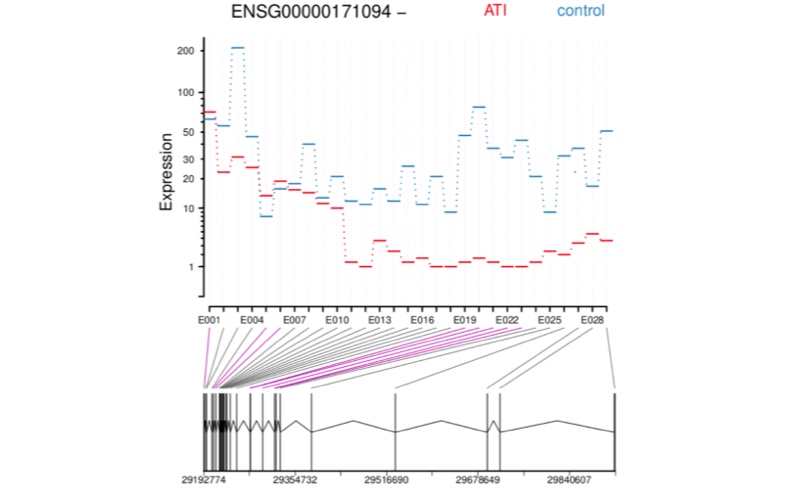
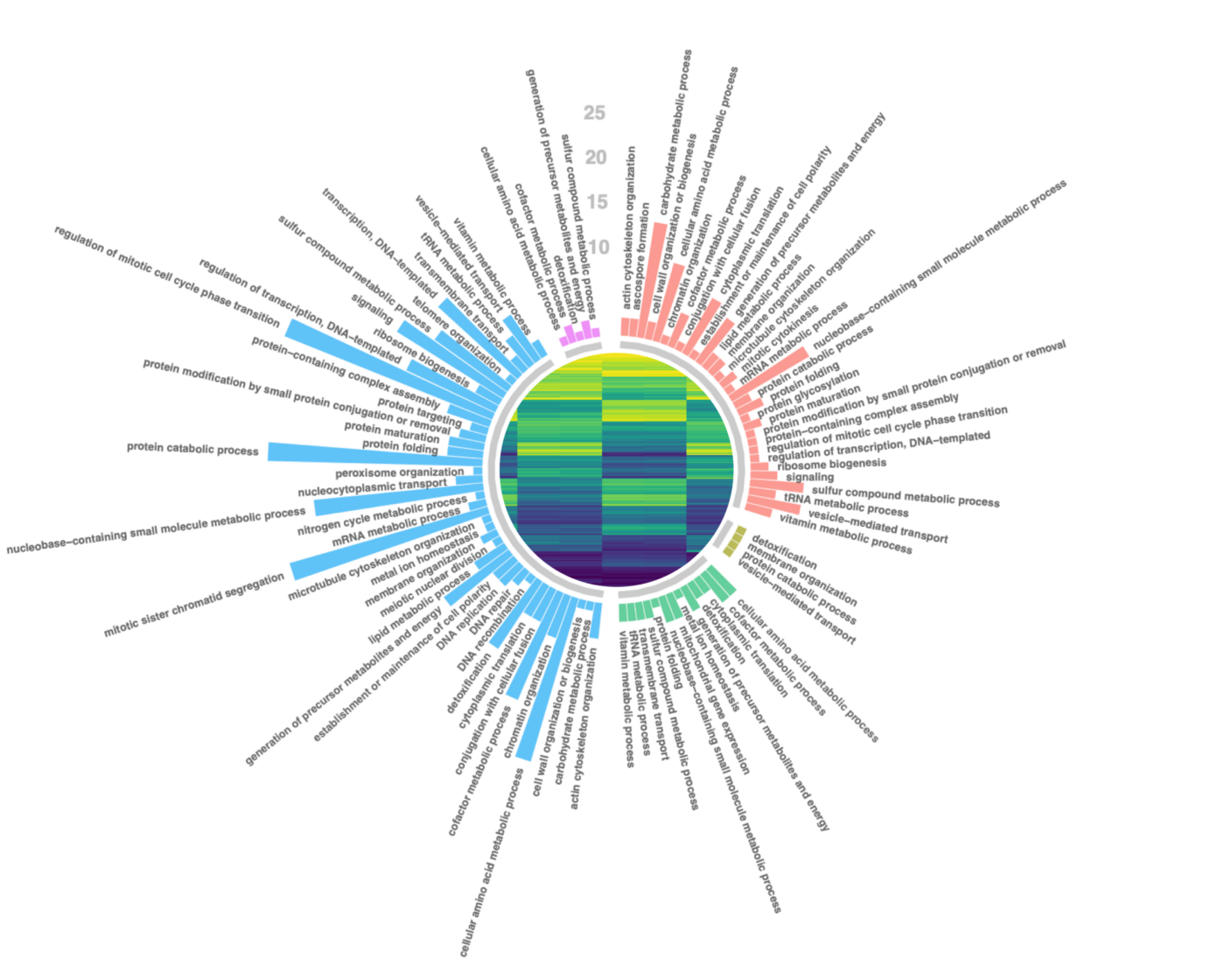
RNA-Seq Structural alteration analysis
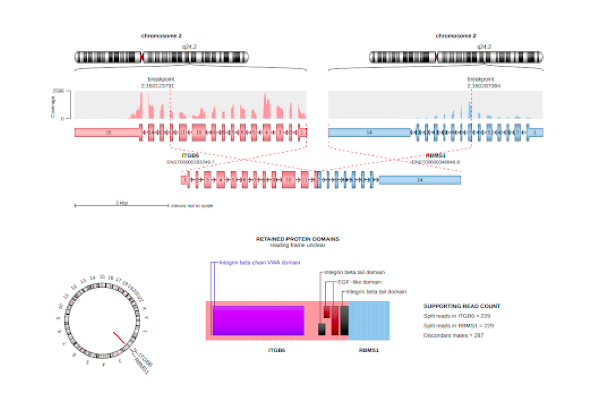
RNA sequencing (RNA-Seq) represents the most precise approach for the characterization of transcript isoforms both known and novel. Compared to previous techniques, RNA-Seq does not require designing probes for selected candidate genes, making it an effective method to explore the full transcriptome and reveal novel transcripts, alternatively spliced genes and other structural rearrangements such as gene fusions.