Shotgun Metagenomics Genome-Resolved Analysis
Shotgun metagenomics offers a unique view into the unculturable microbial community. It enables sequencing an entire microbiome community without culturing restrictions.
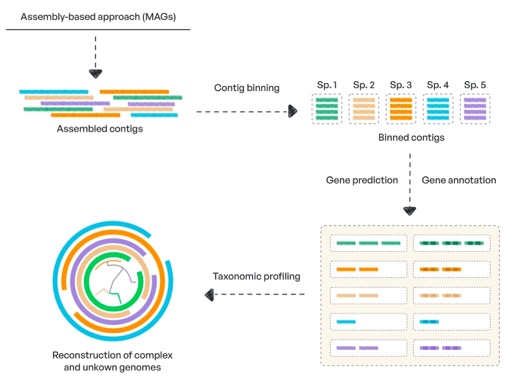
Genome-resolved metagenomics analysis involves the reconstruction of bacterial genomes using shotgun reads. It enables the study of previously unknown bacteria in detail. This is critical for discovering new organisms and genes as well as reducing the amount of microbial dark matter.