Shotgun Metagenomics Marker Based Profiling
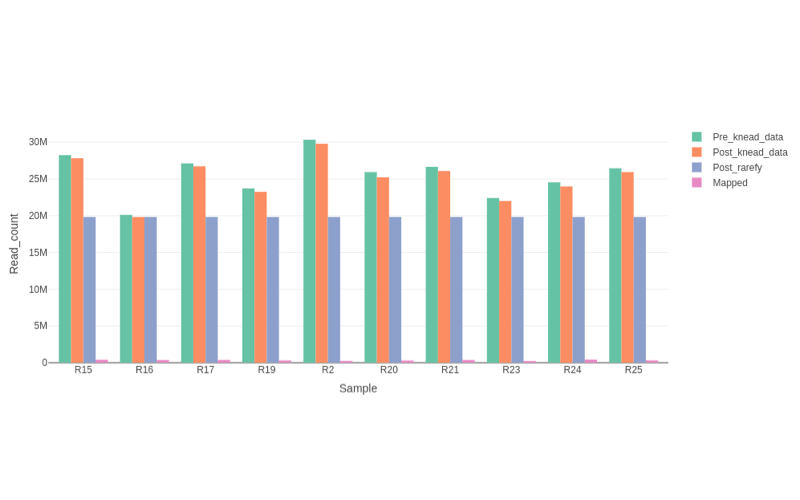
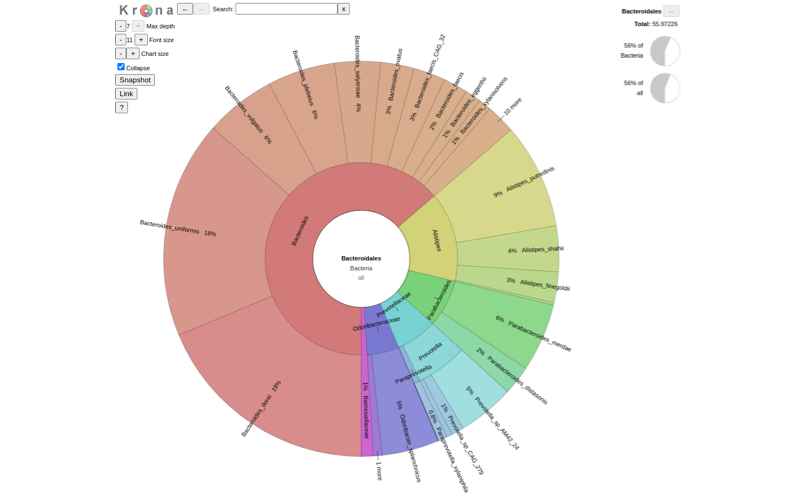
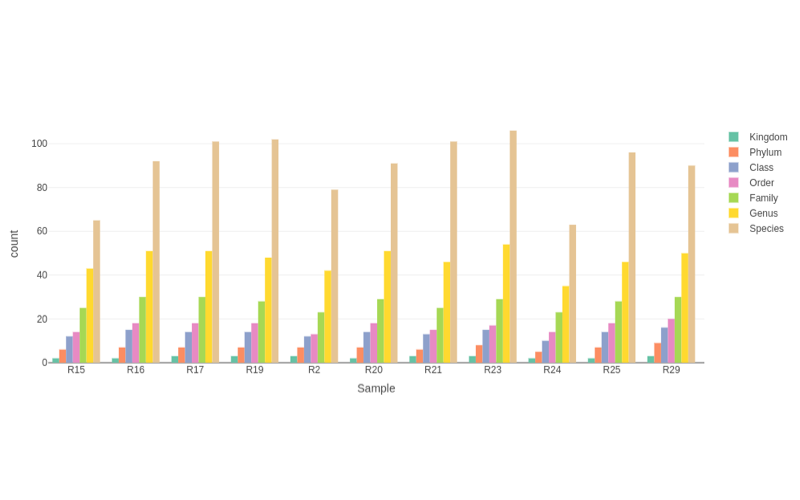
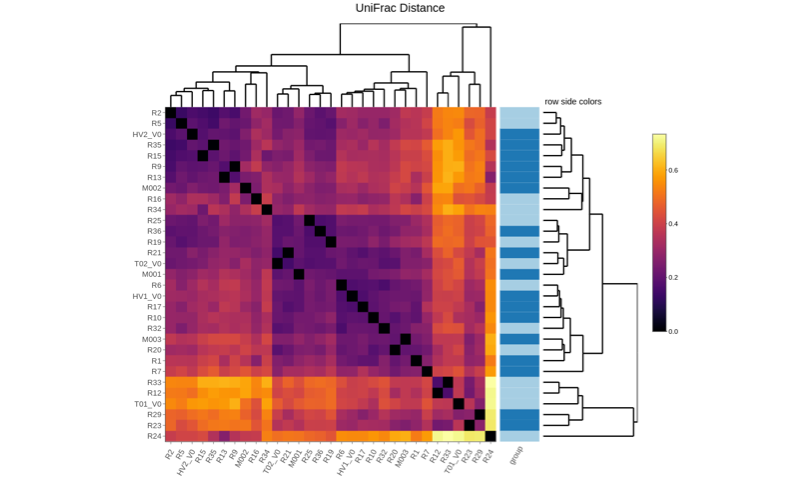
Shotgun metagenomics provides information of the total genomic DNA from all organisms in a sample. Unlike previous techniques, shotgun metagenomics allows investigators to sequence thousands of organisms in parallel from a single sample, obtaining comprehensive information to evaluate bacterial diversity, microbial abundance and compare data from different time points and environments.
The importance of microbiomes has been underappreciated for a long time. With the aid of next-generation sequencing (NGS), the significance of metagenomics and its impact in medicine, ecology, industry and many other areas is now being revealed.